Synthetic Biology & Biophysics
Genome Organization
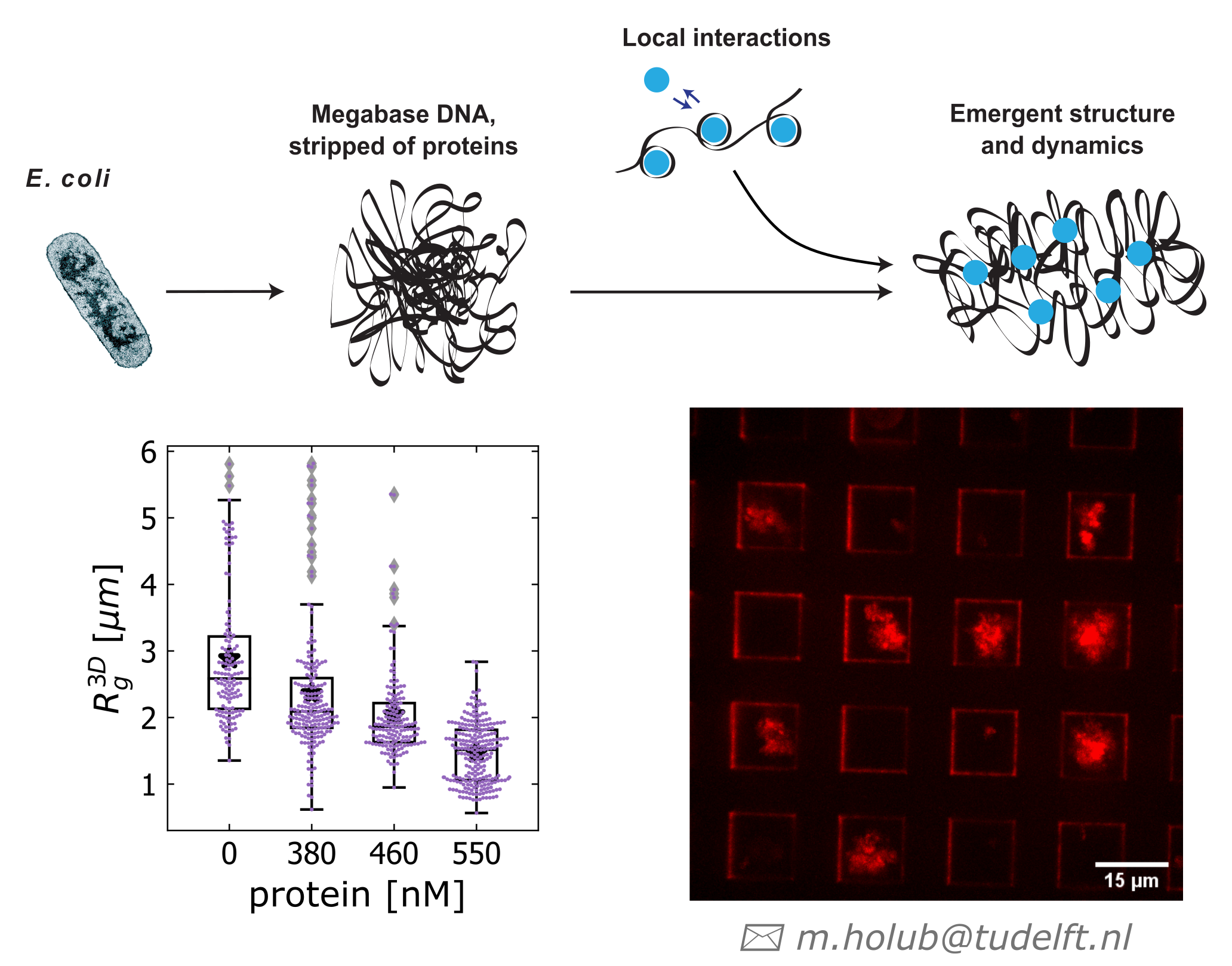
I am currently researching the spatio-temporal organizing principles of megabase chromosomes as a PhD candidate in the group of Cees Dekker at the Department of Bionanoscience at the Kavli Institute of Nanoscience in Delft. I conduct experiments with bacterial DNA in microfabricated compartments that shed light on how tiny proteins and enzymes control its organization and how this, in turn, results in patterns of gene expression. By understanding different ways how a particular protein influences expression of genes, we gain information that can be used to for example develop new antibiotic compounds and build functional synthetic chromosomes from the bottom up.
We have various open student projects for bachelor and master students (including exchange) who want to do experimental wotk at the interface of biophysics, molecular biology, synthetic biology, and microfludics and nanoscience. We can further offer image analysis projects for students with previous experience in Python. Please drop me an e-mail if you are interested, I am happy to discuss and tailor project to your interests!

iGEM and Synthetic Biology
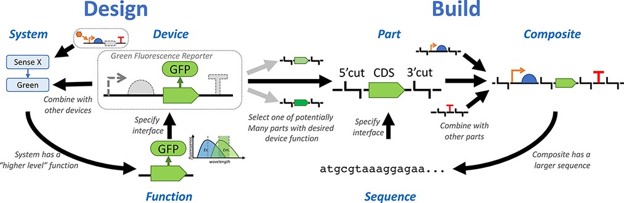
I am also involved with iGEM as a supervisor and judge, and previously published with some folks on why it is important to focus the design of biological systems on function, rather than on sequence.
publications
journals
- Holub, M.; Birnie, A.; Japaridze, A.; der Torre, J. van; Ridder, M. den; de Ram, C.; Pabst, M.; Dekker, C. Extracting and Characterizing Protein-Free Megabase-Pair DNA for in Vitro Experiments. Cell Reports Methods 2022, 100366. https://doi.org/10.1016/j.crmeth.2022.100366.
- Aldulijan, I.; Beal, J.; Billerbeck, S.; Bouffard, J.; Chambonnier, G.; Ntelkis, N.; Guerreiro, I.; Holub, M.; Ross, P.; Selvarajah, V.; Sprent, N.; Vidal, G.; Vignoni, A. Functional Synthetic Biology. Synthetic Biology 2023, 8 (1), ysad006. https://doi.org/10.1093/synbio/ysad006.